Sitemap
A list of all the posts and pages found on the site. For you robots out there, there is an XML version available for digesting as well.
Pages
About
About me Read more
Archive Layout with Content
Posts by Category
Posts by Collection
CV
Cirriculum Vitae
My personal hobbies
Markdown
News and blog-style posts
Page not in menu
Page Archive
The Nordquist family
Portfolio
Publications
Research projects and techniques
Sitemap
Posts by Tags
Teaching Experience
Blog posts
Jupyter notebook markdown generator
Posts
Postdoc Service Award

I was awarded the Postdoctoral Fellow Excellence in Service Award. It was a great honor to be recognized by the University for all the work I do with outreach to local students. I love interacting with student of all ages, including young kids at an annual halloween event, middle-school kids with a mix of youthful energy and a sense of responsibility for important questions, and high-schoolers who are starting to imagine their role in society and solving hard problems. Read more
UMB Ramp Lunch & Learn

I volunteered at the UMB RAMP program’s May Lunch & Learn activity, where a UMB researcher meets with a small group of high school students to discuss their research, career development and trajectory, and then give a demonstration with a hands-on activity. The students played a short game of Monte Carlo pi estimation in person, then we wrote some python code to do the same thing. While not directly related to molecular simulation, it relates a few key ideas, like how seemingly “random” events can converge on a stable value, or how useful computers can be to iterate over a simple, repeatable arithmetic task. Read more
Best of Biophysical Journal 2023!

My paper on hydrophobic dewetting, published with Jianhan and Zhiguang, was selected as one of twenty articles in the Best of Biophysical Journal 2023! This is a big honor, and feels immensely gratifying! This was essentially my first attempt to guide the direction of a research project more directly, although Jianhan of course provided many mission-critical suggestions and corrections at every phase of the project. The honor came as a surprise since none of us knew about the honor until seeing the issue in print after the BPS Annual Meeting in Philadelphia. Check it out! Read more
PhD Dissertation Defense

I gave my final PhD dissertation defense on Monday, June 5th 2023. This was a very special day, and week, to reflect on the work I’ve done and the progress I’ve made as a scientist, and to share that progress with my family, friends, and the academic community here in Amherst. It’s been a wonderful and challenging expeirence. Read more
Postdoc at UMB
I have accepted a position at the University of Maryland, Baltimore, in Alex MacKerell’s lab in the School of Pharmacy. In addition, I have been awarded funding through the Cancer Biology T-32 training grant. I will utilize simulation techniques devleoped by Alex’s group to identify drug-like small molecules for targeting ion channels (BK in particular) in cancer cells. 
Read more
1st class done!
My first class, a first-year seminar called “Order and Chaos, Reconciling atomic randomness and the human world” was a big success. Some of the final projects students designed, carried out and presented:
- Gas permeation through a eggshell-like membrane (pictured below)
- Irreversibility of an explosion simulation
- Thermal equilibration and planet formation
- Equilibration of plucked strings
ResearchFest Presentation

For my recent presentation at the UMass Chemistry ResearchFest, I was awarded the second-place award, a great honor and a generous monetary prize. I was among 4 student speakers selected for the highly-competitive presentations each year. One senior student per lab can be nominated. I was following in the tradition of the great Xiaorong Liu, who spoke at my first ResearchFest in 2018. Read more
Eureka! girls science camp
CNS Teaching Fellowship

The UMass College of Natural Sciences offers a First-year seminar to CNS-declared majors under the broad umbrella “Think like a Scientist”. Graduate students interested in a teaching career are encouraged to apply. Each FYS is unqiue to the person teaching it, which gave me the opportunity to build a (mini) cirriculum and practice some skills I learned abstractly by taking CIRTL online training courses. Read more
Introduction to Evidence-based Teaching
This online course (through the edX platform and CIRTL) introduced me to the kinds of challenges facing an ‘expert’ in a field who is trying to reach a collection of students with various skills, previous knowledge, and emotions. In fact, the growth-mindset and iterative, active learning priciples I hope to implement are the same I need to use to learn how to teach. 
Read more
The Inclusive STEM Teaching Project Course
This online course (through the edX platform) used embodied case study videos and self-reflection prompts to train grad students, postdocs and current faculty to approach education through personal and inclusive strategies backed by the latest educational reserach. 
Read more
Erik and Zoes wedding
Erik and Zoe got married in Eagle, ID. We met at the College of Idaho in Caldwell, ID. 
Read more
Awarded CBI Fellowship
The Chemistry Biology Interface (CBI) is a competitive NIH-funded fellowship designed “to train students with diverse scientific backgrounds for productive research at the interface between chemistry and biology” at UMass.” 
Read more
Website built
My colleague Xiping Gong and I learned that one can host a free website at username.github.io using a special repo with that name. He then found the awesome template academicpages.github.io, which I immediately forked and started using. Since then, many of my colleagues have followed suit: Read more
hobbies
people
publications
research
Biophysics and Machine Learning
It is often useful to leverage computational biophysical data and machine learning to fill gaps in experiments. I’ve used this approach with the chaperone Hsp70, BK channel, and to predict druggable sites. 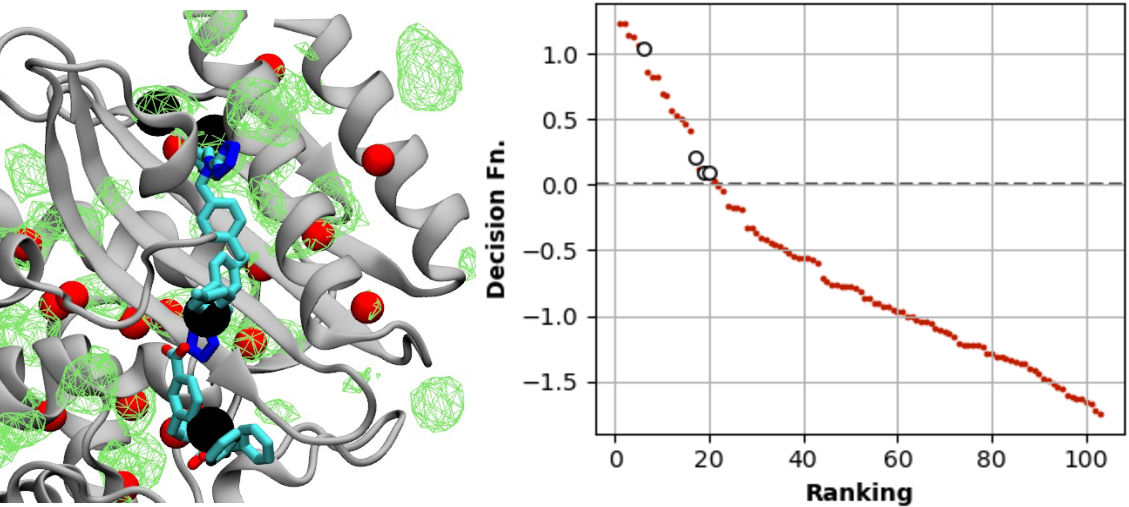
Read more
Hydrophobic Dewetting in protein ion channels and model nanopores
Hydrophobic dewetting forms a vapor barrier and closes BK Channel; how do mutations or drug-like molecules (de-)stabilize this delicate liquid-vapor interplay?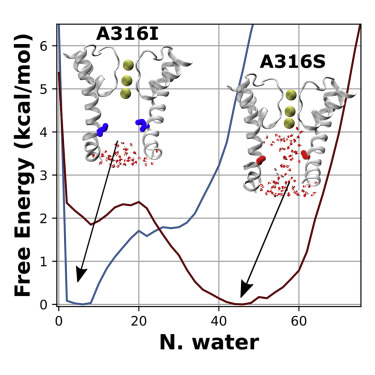
Read more
Computer-aided drug discovery
Using simulations of proteins and small molecules to identify new drug candidates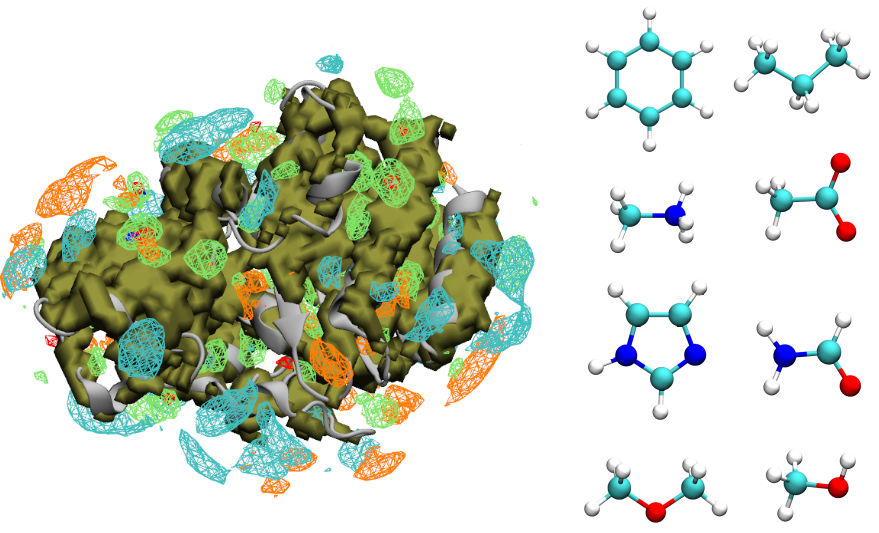
Read more
Molecular dynamics simulations
Using Newton’s laws to simulate molecular motion, like these wiggling waters:
Read more
Computational design of cancer drugs with SILCS
PROTACs are powerful ways to expand the druggable proteome, critical in cancer and other challenging diseases, but their design is often slow and expensive. Hence, CADD is useful!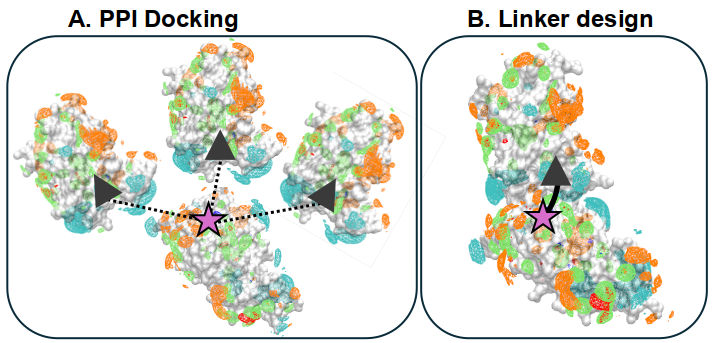
Read more
teaching
Undergraduate Research Mentoring
Training Courses
I have taken several courses associated with CIRTL on the online learning platform edX, including:
Read moreScientific Storytelling
Effective scientific storytelling is similar to telling a fairy-tale.

Read more
Natural Sciences First-Year Seminar
The goal of the FYS is to welcome first-year CNS students and prepare them for college and for a major in the sciences. Read more
Guest lectures
- Molecular mechanics and additive protein force fields; UMass Chem 586 Statistical Mechanics
- Feb 2020, Instructor: Dr. Jianhan Chen
- (Guest moderator) Student-led discussion on AlphaFold2 Nature paper; Amherst College Biophysics Seminar
- Mar 2023, Instructor: Dr. Ashley Carter
- Drug design and CADD in Cancer Biology; UMB Grad. Program in Life Science 665 Cancer Biology: From Basic Research to the Clinic
- Sep 2023, Instructor: Dr. Rena Lapidus.
wiki
00readme
The name of this file follows a convention in the field to put critical overview and introductory information into a file called “README” or “00readme.” The leading zeros help to keep the file sorted at the top if sorting files alphabetically. Read more
Text editors
Text editors are where you will spend the majority of your time, writing scripts for bash/slurm to run simulations, editing simulation input parameter files, writing analysis scripts in python, etc. You will want to get comfortable using one which helps you to be productive! For quick, one-off tasks, the GNOME Text Editor gedit works fine. In the long run, you will want something with more advanced features and integration into bash. NOTE: Don’t use LibreOffice or MS Word! Read more
vi - VIsual editor
My shell-based visual editor of choice is vi. It is a very powerful and ubiquitous tool, but involves an initial learning curve. The content of this entire website was written/edited inside of vi. Note that you can save your preferred configuration in the user config file ~/.vimrc. Read more
GNUplot
It is often highly convenient to make a quick plot of a function or data right from the command line. The gnuplot utility provides this. If desired, one can make high-quality plots as well. However, I recommend using python for plots made regularly or for serious presentations or publications, and using gnuplot for on-the-spot analysis. Read more
Bash
Bash, or the Born-again shell, is one of the most popular and therefore well-documented shell environments, making it an ideal choice for beginners and experts alike. It is the default on many Linux distributions, and was in MacOS until 2019 (Catalina) when the default became z-shell. Note that the Windows shell Command Prompt (cmd.exe) is not Unix-based. Read more
Shell Keybindings
On most shells, including bash, there is a set of incredibly simple and powerful keyboard shortcuts that you should learn and get comfortable using. I’ve copied this cheatsheet from here: Read more
Bash commands
The Unix programming paradigm is built around small programs that read and write plain text in simple, predictable ways. By chaining tools together with redirection and pipes, complex workflows become compact and expressive. This approach is powerful for quickly processing large text files like logs, or large text-based datasets, all right at the command line. Read more
Bash scripting
The Unix programming paradigm is built around small programs that read and write plain text in simple, predictable ways. By chaining tools together with redirection and pipes, complex workflows become compact and expressive. This approach is powerful for quickly processing large text files like logs, or large text-based datasets, all right at the command line. Read more
Looping
The Unix programming paradigm can also allow for programming technqiues like looping, which allows for more complex scripts that extend beyond a single line. Read more
More
While there are always more commands one can learn, I’ve tried to give a list of some very commonly used ones which are not quite as urgent to learn for beginners, but essential for proficiency. Let me know if more should be added. Read more
The Slurm Scheduler
On shared computing resources, which can range from a small network to a high-performance computing cluster (HPCC), a scheduler is the critical tool which allows many users to submit jobs simultaneously and schedule which jobs get assigned to which resources. Schedulers are powerful and complex tools, but with a few key ideas you can get started. Read more
Unix filesystem
Bash, or the born again shell, is a very popular and well-documented command line shell environment for working in the Unix filesystem, and consists of directories (like folders) and files. There are many good overviews to the shell environment I prefer: bash or the born-again shell. The internet is full of great tutorials on unix and bash commands and scripting. Read more
Wiki
Hello, and welcome.
Read moreCHARMM-GUI
There are few tools which have democratized molecular mechanics simulations more than CHARMM-GUI. It is an extremely widely-used tool for setting up simulations for a wide variety of systems (biological and materials), including fairly non-trivial systems and enhanced sampling setups. Input scripts including force fields are automatically generated. It is a powerful first step in setting up many simluations, especially for beginners. Take time to play with settings and appreciate some of the (admittedly few) limitations. Read more
Engines
The MD engine is the sofware that calculates the forces and propagates the atoms, consisting of tools for force integrators, thermostats, barostats, etc. Modern software is increasingly complicated as there must be a separate version complied to run partially or entirely on a GPU. There are many major MD engine software, of which I have primarily used CHARMM, Open-MM, and GROMACS. Other major interfaces include Amber, NAMD, LAMMPS, etc. Historically, most/all also were developed alongside a force field with the same or a related name. Although perhaps not 100% true in all cases, you can largely run most force fields on most engines today with the correct setup. Read more
Force Fields
The MD Force Field is the term used to refer to what the equation used to obtain forces acting on particles in molecular mechanics simulations. Often, the fundamental equation written down in papers is a Potential Energy equation (below), where the forces are obtained by the first derivative. In this page I only list all-atom additive force fields, although many other MM force fields exist (polarizable, coarse-grained, etc). A critical component is the so-called “general” force field, which refers to a set of parameters and/or method for generating parameters for arbitrary small molecules such as drugs. Read more
Molecular Mechanics
Quantum mechanics is the foundation of modern chemistry and, by extension, all of computational chemistry. It was developed in the 1920s to address several outstanding challenges, including explaining atomic spectra and the wave–particle duality of electrons. A central simplification enabling practical calculations is the Born–Oppenheimer approximation, which separates nuclear and electronic degrees of freedom by exploiting their vastly different timescales. While this makes molecular quantum calculations tractable in principle, the computational cost still grows steeply with system size. Even with modern methods, fully QM simulations of systems (ab initio MD) are limited to at most ~100 atoms and timescales on the order of picoseconds. Read more
Molecular Dynamics simulations
This is a huge topic area… best exploring by reading and discussing seminal and current research publications, although there are textbooks available which cover key topics. Read more
Reading list
The core topics in the field of molecular mechanics I tend to think of as belonging to two core subtopics: the MM calculations, and the force fields. The MM calculations are performed, sophisticated software “engines” that are responsible for calculating the forces acting on many atoms and propogating their positions forward in time. The force calculation is based on a simple equation (“force field”) that describes how the force acting on a given atom is a function of it’s position with respect to other nearby atoms. Read more
ATP Synthase
ATP synthase is a critical enzyme in cellular energy production. It converts ADP to ATP using a proton gradient across membranes. ATP, often referred to as the energy molecule, stores energy by compressing many negatively charged phosphates together, connected by high-energy phosphoanhydride bonds (P-O-P). By breaking the bond, energy can be released and coupled to many other unfavorable processes. This is a key part of how life can exist so far from thermodynamic equilibrium! The animation below shows the rotary mechanism of the synthase: Read more
Some cool stuff
This is a small, but growing collection of some of the many proteins that I find cool. Read more
Kinases
Kinases are enzymes that catalyze the transfer of a phosphate group onto a substrate protein. Phosphorylation changes their activity, localization, stability, an/or interactions. Protein kinases form one of the largest enzyme families in eukaryotes and are central regulators of nearly all cellular processes. The enzymatic activity depends on the kinase, but often they recognize specific sequences of amino acids and modify one or more of Tyr, Thr, and Ser. Although different detailed mechanisms exist, in general kinases stabilize a high-energy intermediate such as through the presence of positively-charged residues nearby. Read more
TRPV1
Transient Receptor Potential 1, a.k.a. “the spicy sensor”
The 2021 Nobel Prize in Medicine was awarded to the scientists who discovered that some protein channels are responsible for our sense perception. Below, observe the TRPV1 channel, bound to the “spicy” flavor molecule capsaicin. Along with the other TRPV channels, TRPV1 is responsible for sensing temperature changes. Interestingly, they discovered that when capsaicin molecules bind to TRPV1, it activates in the same way as when our bodies get warmer. When we eat spicy food, our brains recieve similar chemical signals as when we get hot. These discoveries helped to explain how our brains, which are fundamentally chemical machines, interact with the external world. Read moreProteins
Proteins are remarkable molecules, critical for life in many interconnected ways. They perform many critical functions inside of living cells. A particularly important example of a critcal function is an enzyme, or a catalyst. A classic enzyme is a kinase, which will be discussed in detail on it’s own page. Read more
Protein sequence
Proteins are made of sequences of amino acids which condense into complex polymers, forming peptide bonds between the amine and carboxylic acid groups of separate amino acids, and shedding a water. The leftover group is called an amino acid residue, sometimes informally just called a “residue.” Note that residues can be grouped into various loose categories based on the chemical group (“sidechain”) which is coming off the the core “backbone” (N, C\(\alpha\), C=O). The degree of hydrophilicity of these sidechain groups is critical. In the salt water environment of the cell, entropic and enthalpic interactions drive the overall chain to fold such that hydrophobic groups get buried and hydrophilic groups get exposed. By convention, we think of the sequence as being oriented from the amine-terminal (N-term) to the carbonyl-terminal (C-term) end, since is also how the protein is synthesized in the ribosome. Read more
Protein structures
Proteins fold spontaneously into complex 3-dimensional structures in salt water at room and body temperatures, and they are not only critical for life (and mis-folding critical causes of disease), but also stunningly beautiful to behold! You can spend hours on the World Wide Protein Data Bank (wwPDB), looking at various structures. Play around with these below! You can rotate and enlarge the structures. Notice the various secondary structures such as \(\alpha\) helices (purple) and \(\beta\) sheets (yellow), which are formed by hydrogen-bonding between residue’s amide and carbonyl groups. Read more
Python
python was a paradigm-shifting programming language when it was built in the 1990s, and remains widely in use today by serious programming and beginngers alike. Time spent learning the basics is well spent, as the essential skills will translate to other languages. There is almost no programming task which can’t be done well in python. Read more
Vectorization
Efficient computation via vectorization in the numpy library
Note: This is more of an advanced topic, something to be aware of if you are running slow code but not totally necessary. In a previous challenge, you may have created a program to estimate $\pi$ via a simple Monte Carlo simulation. A simple loop in python is not efficient if one wants to evaluate millions or billions of operations! Vectorization implemented in numpy allows more efficient computations to be carried using very efficient linear algebra implementations in programming languages like C or Fortran. Here is an example code for estimating $\pi$ using numpy (installed with mamba install numpy if needed): Read morePlotting data in Python
Plotting with matplotlib
To run this code, you will need to install matplotlib, such as by running mamba install matplotlib. Here is an example script which makes a scatter plot of a data set. The data file is expected to be space-delimited and be organized so that the x-value is the first column, and then all subsequent columns are interpreted as different y-values. You could create such an input text file using awk to parse a more complex file, for example. Here is an example data file:X1 Y1 Y2 Y3
1 1 1 1
2 2 4 8
3 3 9 27
4 4 16 64
5 5 25 125
Getting started in python
Learning Python
I recommend learning python at a website such as learn-python.org, where you can slowly build up your experience with basic elements of programming. You will find common elements with bash programming, because certain foundational elements of programming such as conditional statements and looping are present in any language. Time spend learning python in this way will translate to nearly any other language. Read moreVMD
Visualizing Molecular Dynamics: a very powerful tool, essential for success! You can download the software, or check out the documentation. For quick reference, there are two common ways to load files in vmd. One can load a single system, possibly with multiple files and frames: vmd topology.psf trajectory.dcd Read more
Materials
You can define custom colors and materials inside the after idle block of .vmdrc. Check out the VMD docs e.g. materials and colors. Huge thanks to my friend Jian Huang for pointing this out here! Read more
Save settings and visualization states
You can save your preferred setting for default VMD states in the file .vmdrc. VMD comes with a GUI editor for this file in the Main window under Extensions > VMD Preferences, which is generally the best way to customize the settings. Play around! Read more

