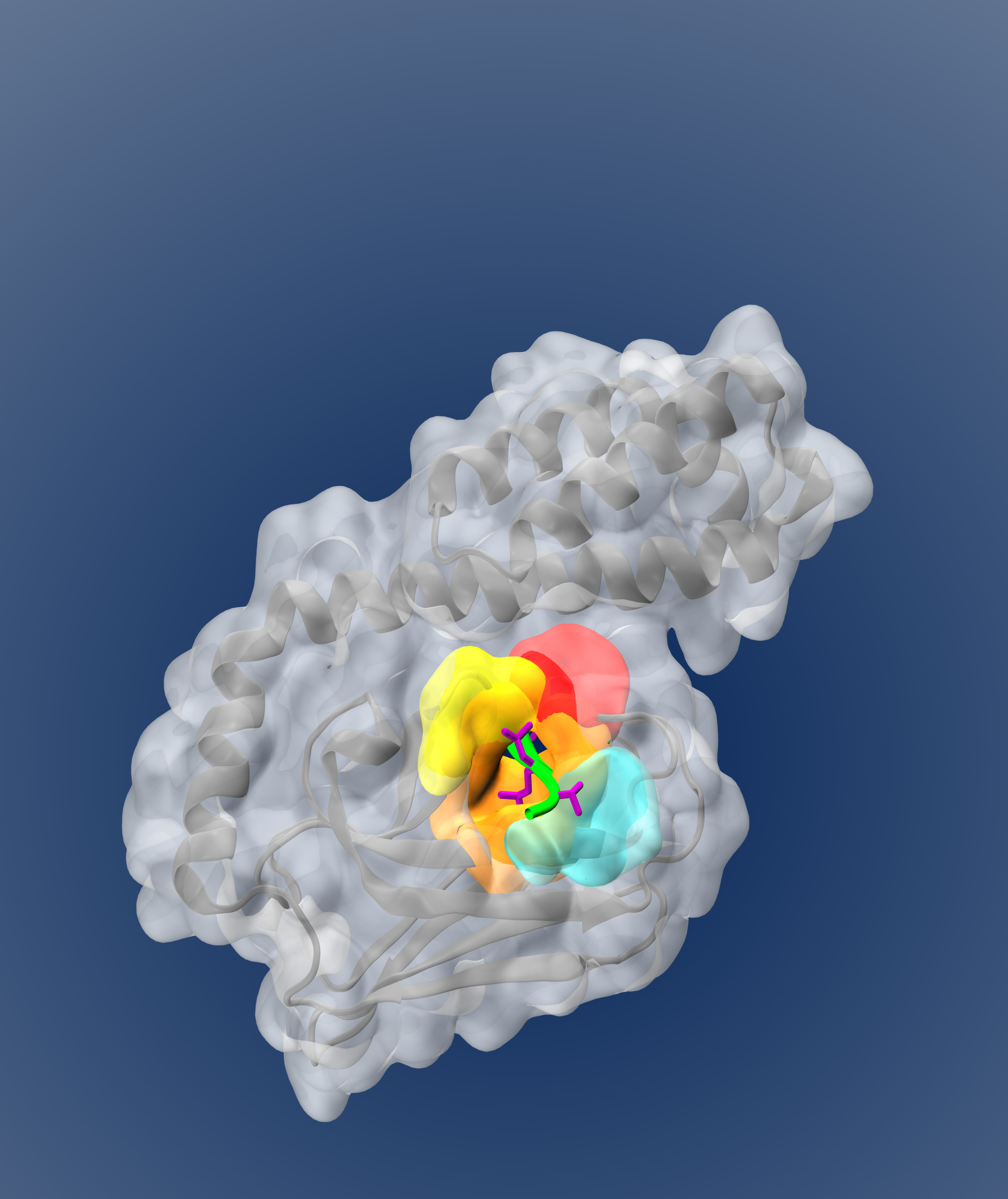
Physics-based modeling of Hsp70-substrate binding
Hsp70s (Heat-shock proteins with molecular weight of 70 kiloDaltons) are a special kind of protein called molecular chaperones and play a central role in the cell's intricate quality control mechanism. They do this in part thanks to a remarkable ability to bind to a wide variety of misfolded proteins without binding to well-folded or intrinsically-disordered proteins. The physical basis for the molecular chaperone Hsp70’s promiscuous selectivity for many substrate peptide sequences, but not all, remains poorly understood. A model explaining Hsp70s selectivity and account for the alternative ‘reverse’ orientation, recently found to be functionally significant, was of great interest to my collaborators in the Gierasch group and others in the field. I first conducted extensive comparative modeling of available structures of substrates bound to DnaK. This structural analysis directly informed an MD simulation protocol for sampling interactions between substrate residues and DnaK. These interactions formed the basis of the model, which I trained to reproduce low-resolution binding fluorescence experiments. I validated the model on the much higher-resolution data set of peptides bound in various crystal structures, where the overall orientation and precise residues at each site are known. The model correctly predicted the preferred interactions and orientation for over 70% of the peptides in crystal structures, which represents the current state-of-the-art accuracy. The model’s physically-motivated terms provide insight into the origins of the selective promiscuity of Hsp70s, and the modeling approach can be extended to problems fitting using lower-resolution data to describe higher-resolution problems. Read the paper, or our review of the subfield or, view the code on Github.