Computational design of cancer drugs with SILCS
Computational design of PROTACs
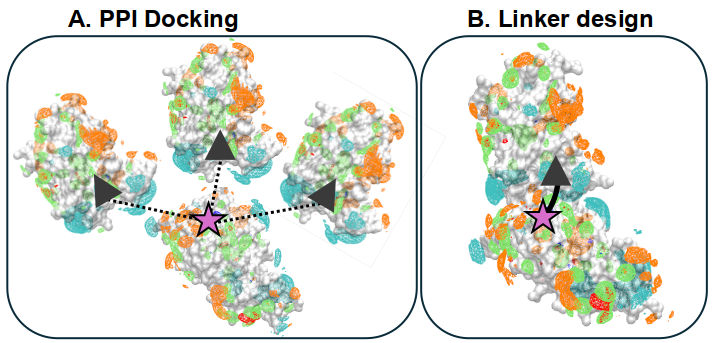
PROTACs, or Proteolysis Targeting Chimeras, are molecules that combine two "warhead" ligands with a flexible linker, designed to increase the probability that the target protein (warhead 1) is degraded by an E3 Ubiquitin ligase (warhead 2), allowing the target protein's to be completely abolished. The PROTAC is catalytic and can have high activity despite transiently binding (due to covalent Ub labeling), enabling a much wider array of targets in principle than available to conventional noncovalent inhibitors. This project is ongoing in the MacKerell lab in Baltimore, MD, and is a part of my training in the Cancer Biology T32 in the Univ. Maryland Cancer Center. The approach will be to leverage the SILCS (GCMC/MD) fragment affinity FragMaps to determine the Target-Ligase Protein-Protein Interactions (PPIs) which can allow a PROTAC to bind, then determine which linkers bind best and presumably stablize the complex. We have constructed a large set of >20 protein-ligase complexes with >700 PROTACS characterized with various biological activity data to guide and benchmark our protocol.